
Teaching
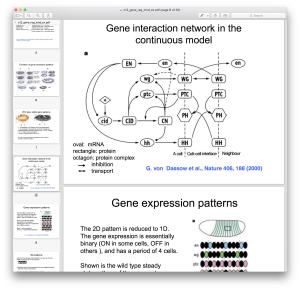
Most complex systems are hard to describe and understand because they are composed of large numbers of elements interacting in a non-ordered way. A good example is cellular biology; diverse cellular components (genes, proteins, enzymes) participate in various reactions and regulatory interactions, forming a robust system.
A very useful representation of complex systems is given by graphs (or networks), where we denote the components with nodes and their interactions by edges. The properties of these interaction graphs can then be analyzed by graph theoretical and statistical mechanics methods and this information can lead to important conclusions about the dynamics of the system.
Courses
PHYS 580: Elements of Network Science and its Applications (graduate)
PHYS/BIOL 465: Network analysis of biological systems (undergraduate)
Lecture Notes
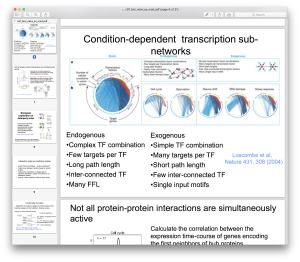
Current lecture notes are distributed via Canvas. The materials covered in the course will change over the years as we incorporate the newest advances in the field. Below we list lecture notes for a few prior years.
-
Lectures notes for 2012: phys580-2012.zip
-
Lectures notes for 2009: phys580-2009.zip
Topics
- Elements of graph theory: node degree, distances between nodes, clustering.
- Node betweenness, subgraphs, directed graphs.
- Random graph theory.
- Network models and theory: lattices, small-world networks,scale-free networks, evolving networks.
- Network robustness and vulnerability.
- Flow processes on networks.
- Introduction to cellular networks: gene regulatory networks, signal transduction networks, metabolic networks; methods of network inference.
- Modeling reaction networks: elements of chemical kinetics.
- Signal transduction network models.
- Modeling gene regulatory networks using continuous and discrete dynamics.
